References
Breda, Jérémie, Mihaela Zavolan, and Erik van Nimwegen. 2021. “Bayesian Inference of Gene Expression States from Single-Cell RNA-Seq Data.” Nature Biotechnology, 1–9.
Hafemeister, Christoph, and Rahul Satija. 2019. “Normalization and Variance Stabilization of Single-Cell RNA-Seq Data Using Regularized Negative Binomial Regression.” Genome Biology 20 (1): 1–15.
Kurtz, David M., Joanne Soo, Lyron Co Ting Keh, Stefan Alig, Jacob J. Chabon, Brian J. Sworder, Andre Schultz, et al. 2021. “Enhanced Detection of Minimal Residual Disease by Targeted Sequencing of Phased Variants in Circulating Tumor DNA.” Nature Biotechnology, July. https://doi.org/10.1038/s41587-021-00981-w.
Love, Michael I, Wolfgang Huber, and Simon Anders. 2014. “Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2.” Genome Biology 15 (12): 1–21.
McCarthy, Davis J, Yunshun Chen, and Gordon K Smyth. 2012. “Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation.” Nucleic Acids Research 40 (10): 4288–97.
Robinson, Mark D, Davis J McCarthy, and Gordon K Smyth. 2010. “edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data.” Bioinformatics 26 (1): 139–40.
Shajii, Ariya, Ibrahim Numanagić, Alexander T Leighton, Haley Greenyer, Saman Amarasinghe, and Bonnie Berger. 2021. “A Python-Based Programming Language for High-Performance Computational Genomics.” Nature Biotechnology, 1–2. https://www.nature.com/articles/s41587-021-00985-6.
Tran, Hoa Thi Nhu, Kok Siong Ang, Marion Chevrier, Xiaomeng Zhang, Nicole Yee Shin Lee, Michelle Goh, and Jinmiao Chen. 2020. “A Benchmark of Batch-Effect Correction Methods for Single-Cell RNA Sequencing Data.” Genome Biology 21 (1): 1–32. https://link.springer.com/article/10.1186/s13059-019-1850-9.
Vallejos, Catalina A, John C Marioni, and Sylvia Richardson. 2015. “BASiCS: Bayesian Analysis of Single-Cell Sequencing Data.” PLoS Computational Biology 11 (6): e1004333.
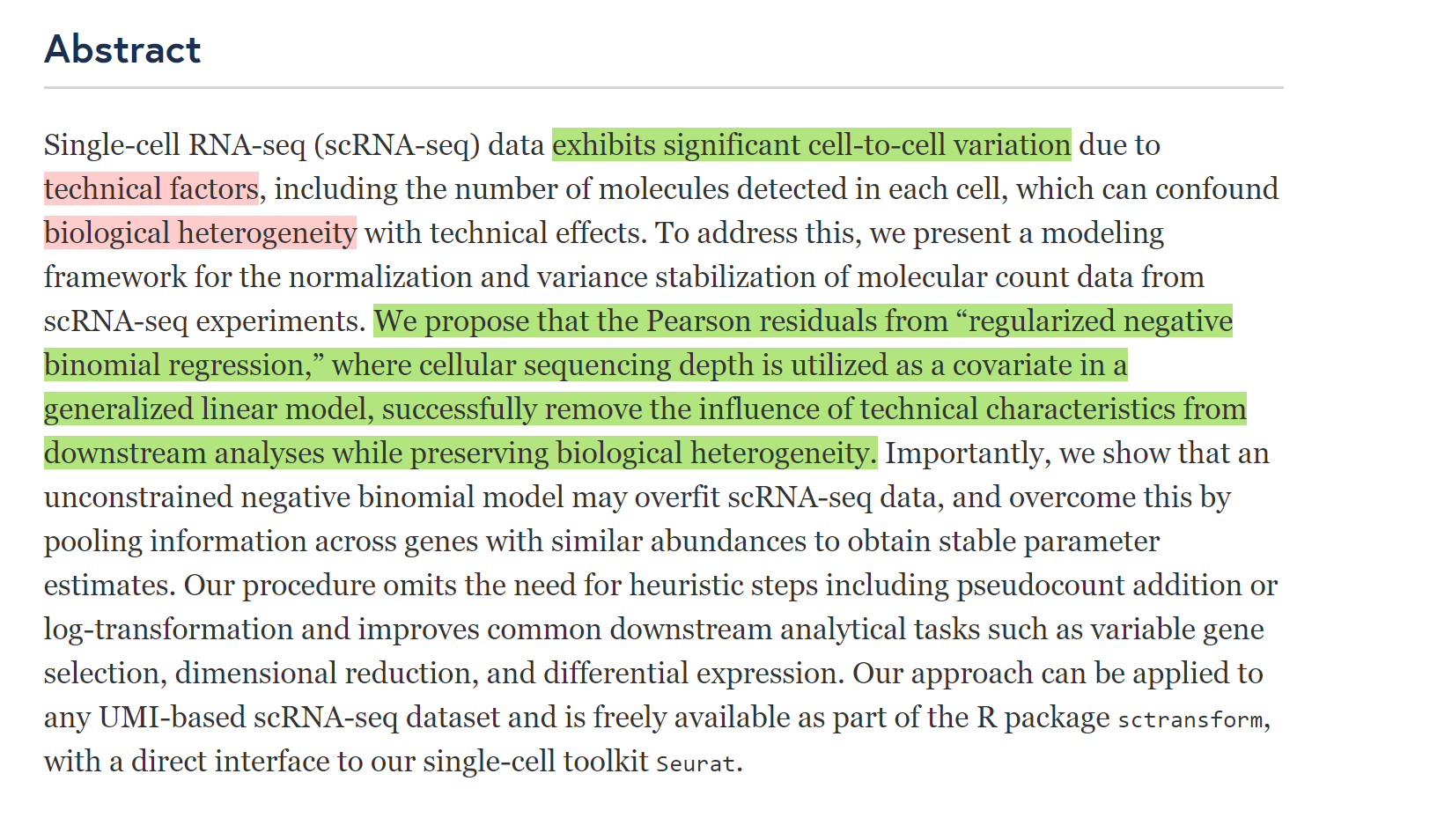


 Results
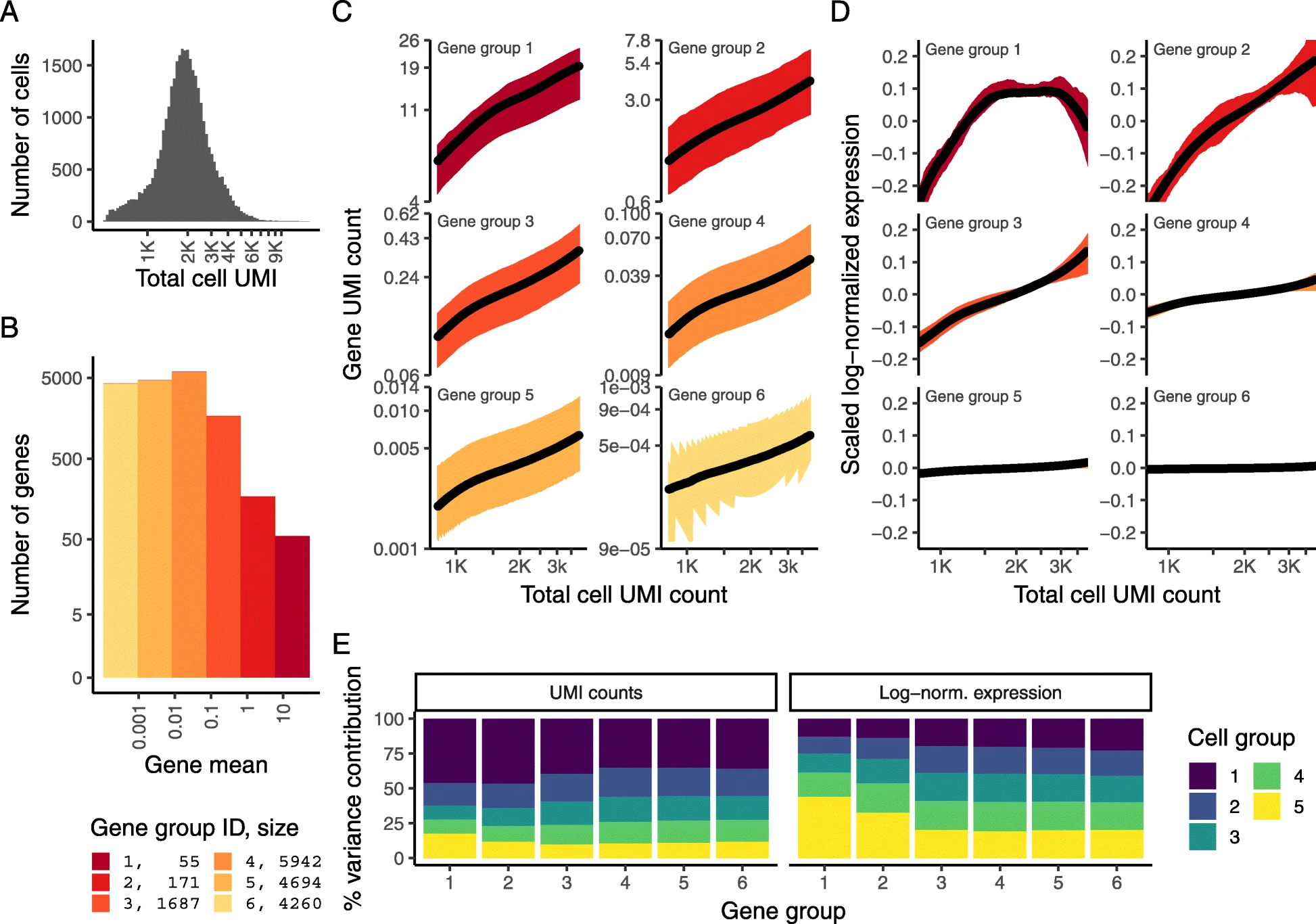
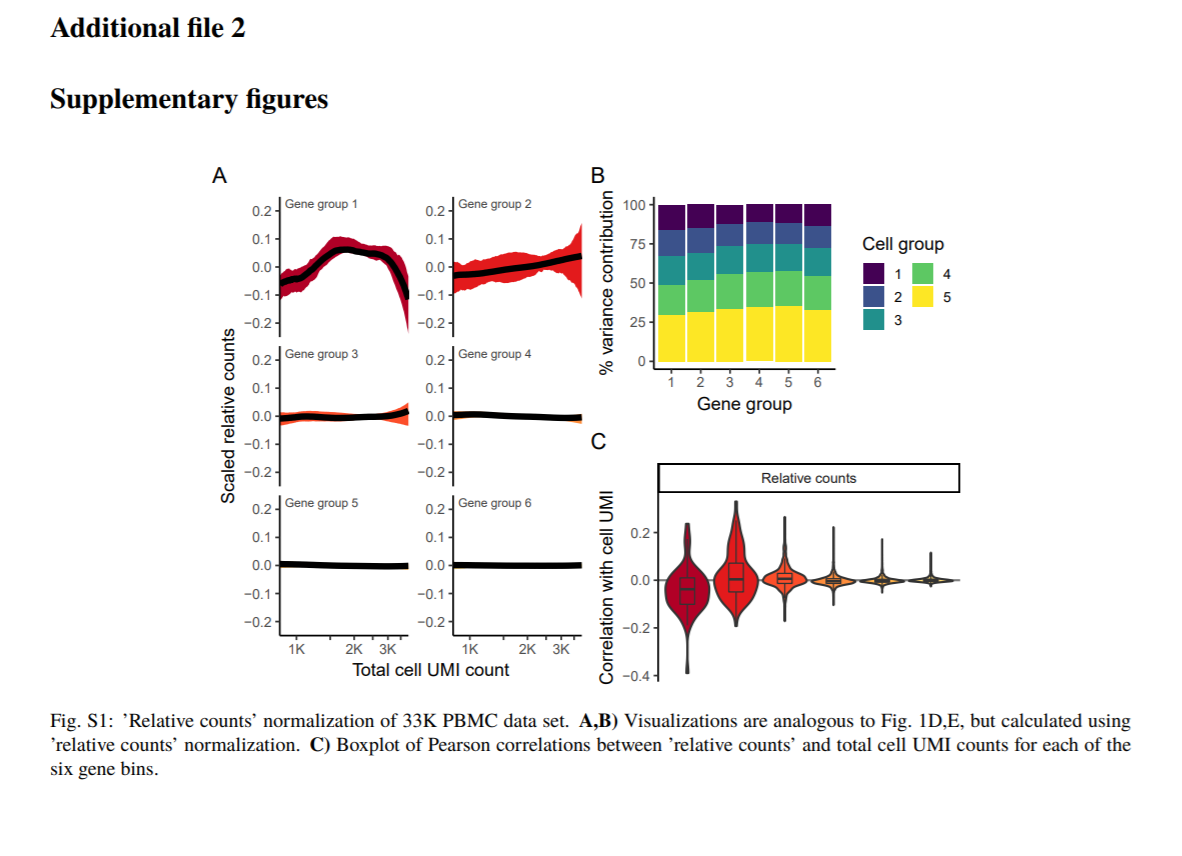
Results
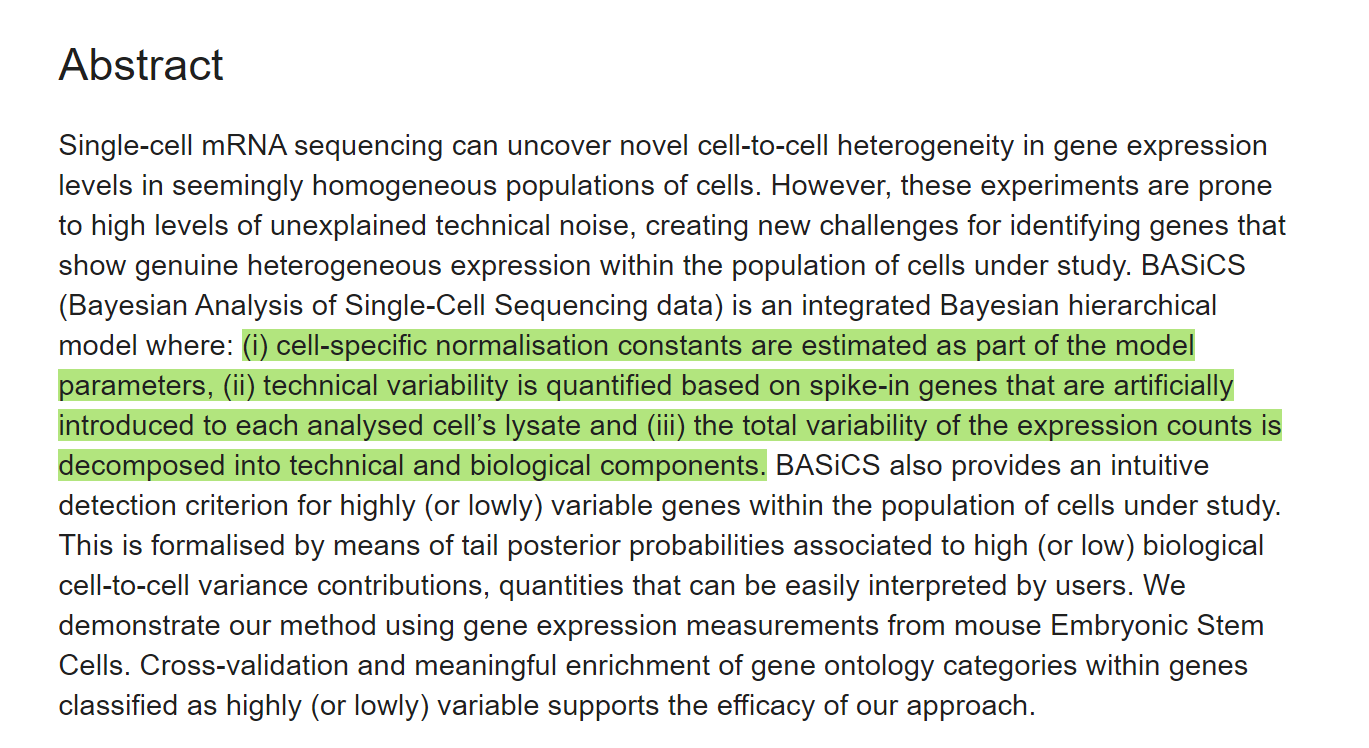
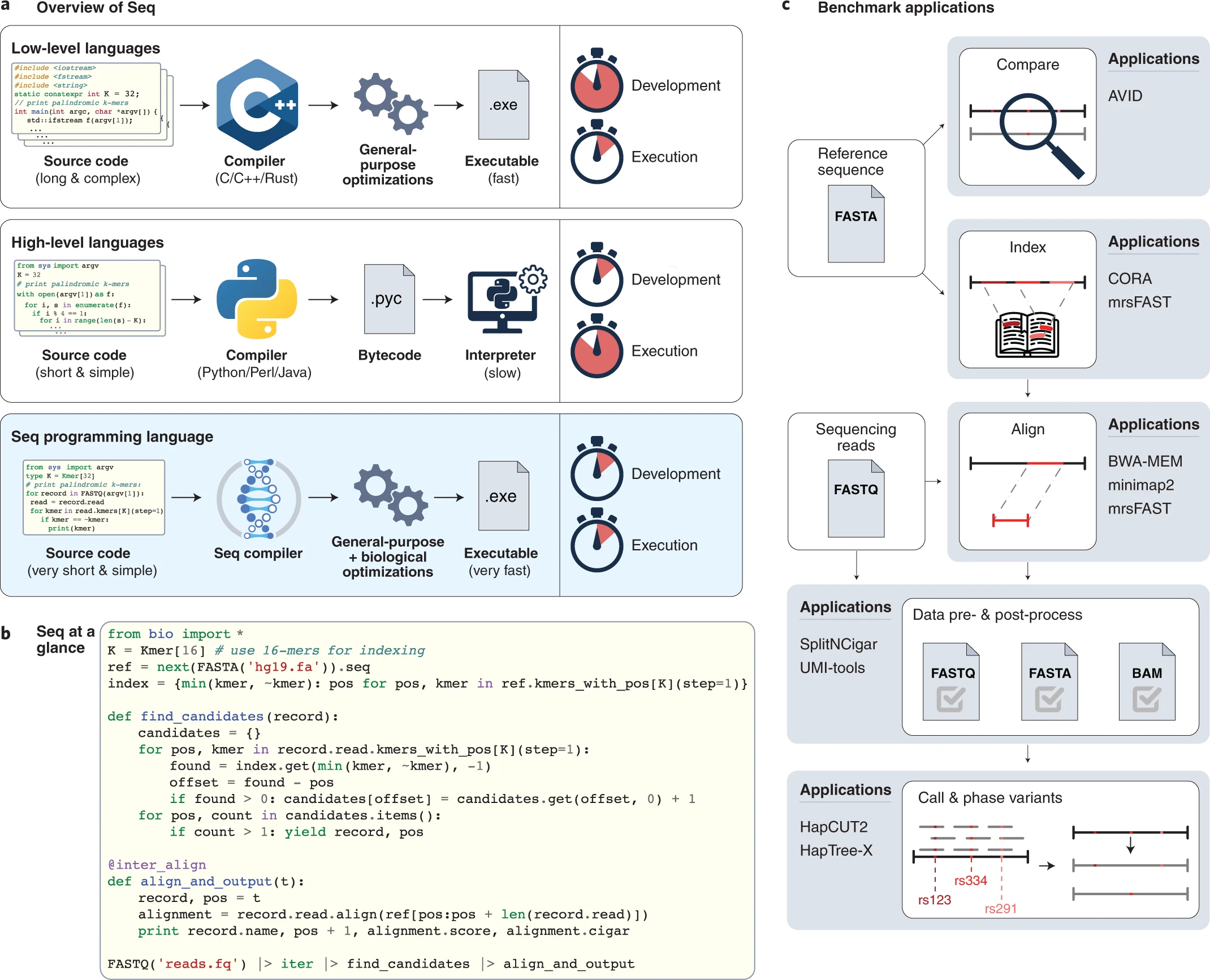
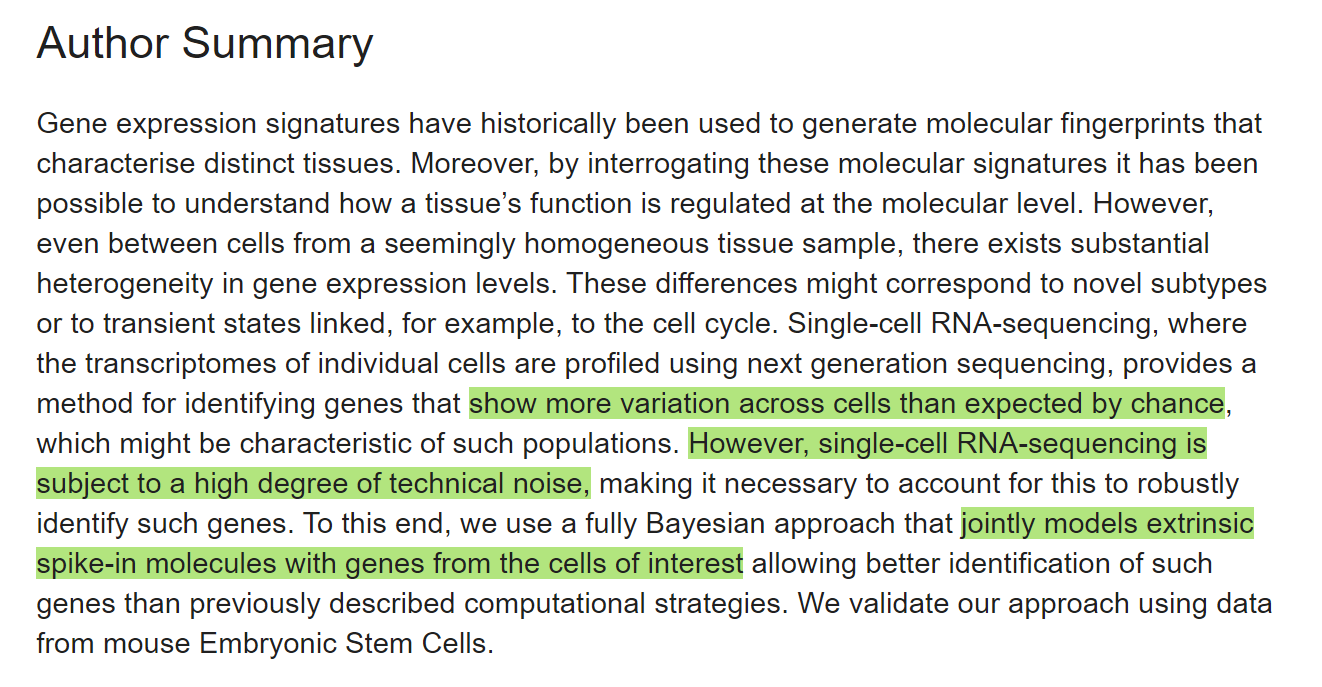 INTRODUCTION
INTRODUCTION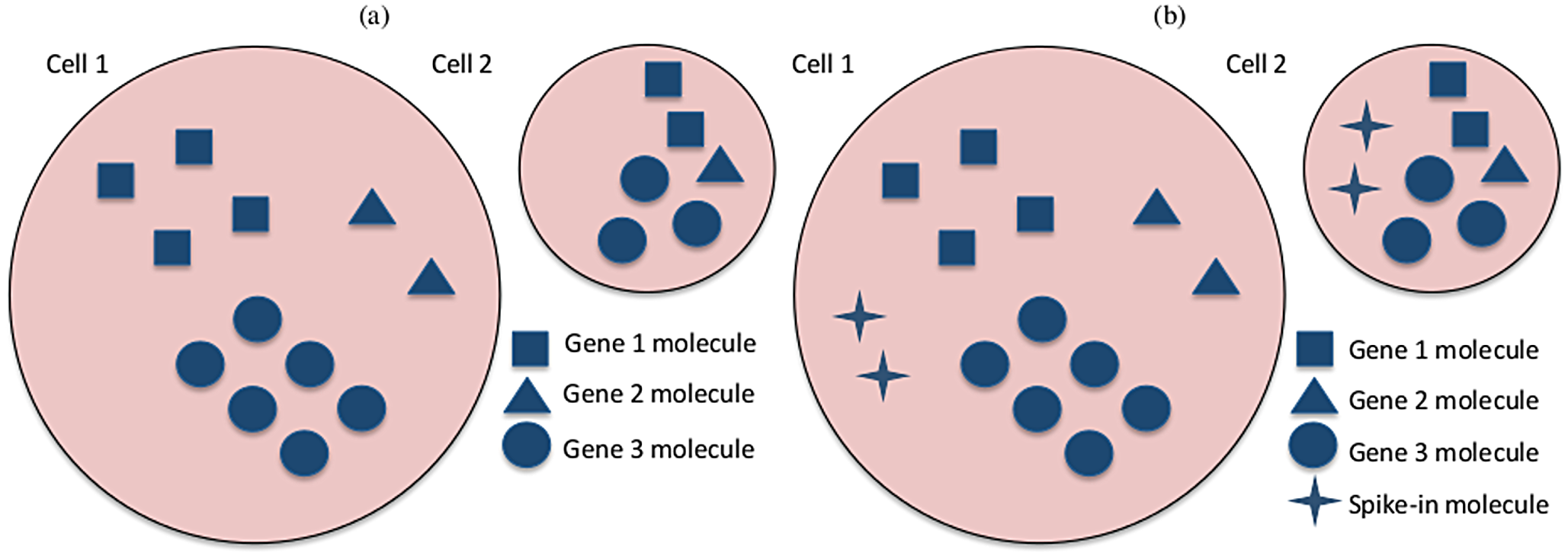
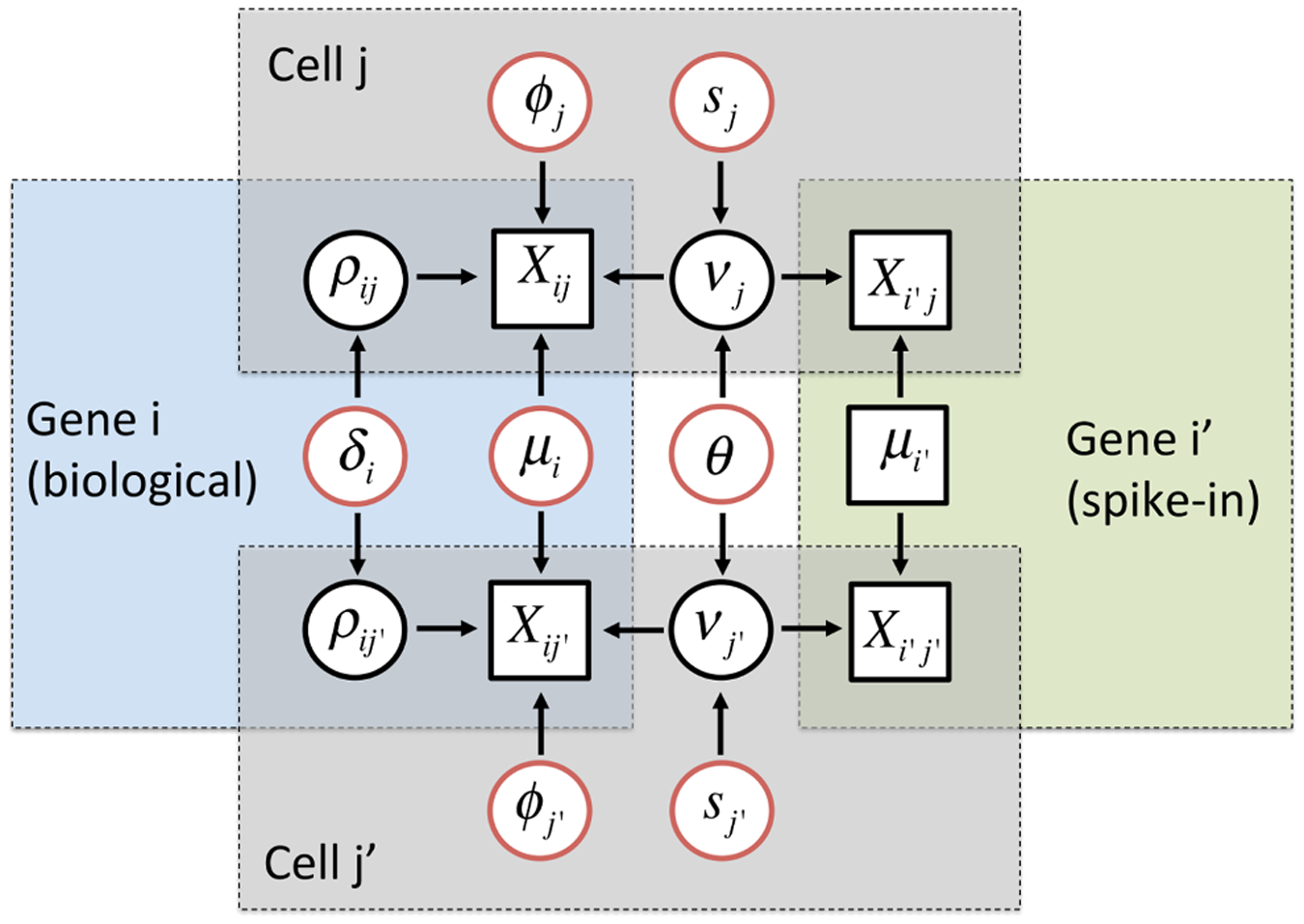
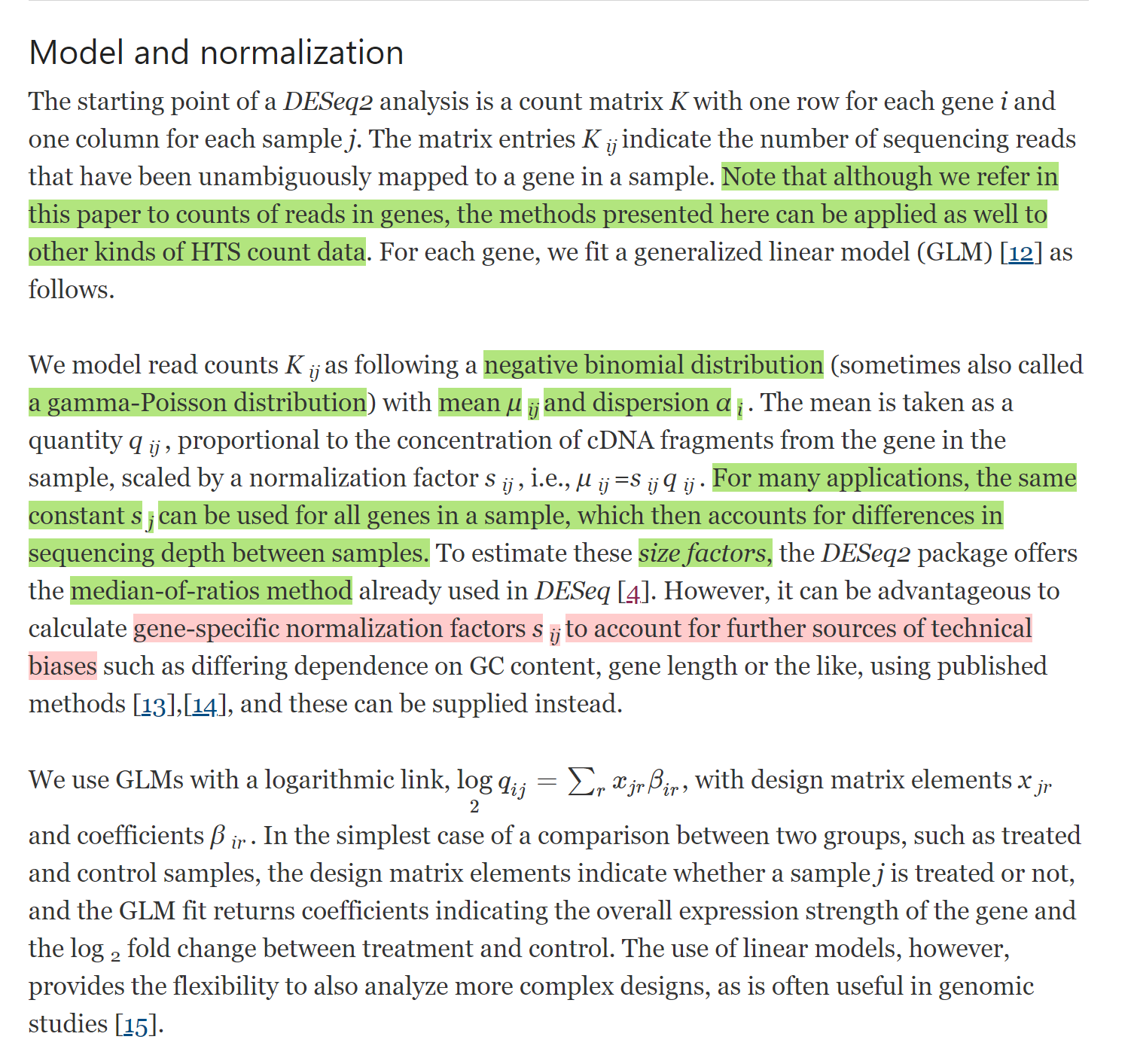
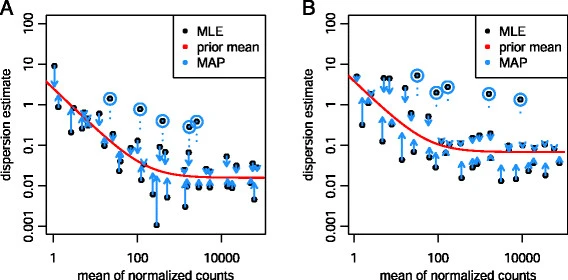
 #### edgeR
#### edgeR